Widespread transcriptional pausing and elongation control at enhancers
02/01/2018
New tool tracks down distant regulators of gene expression, upends expectations 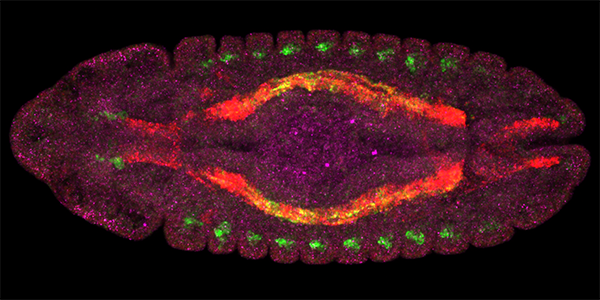 Gene enhancers light up in distinctive patterns in different cell types in a fruit fly.
Image: Olga Mikhaylichenko and Eileen Furlong/European Molecular Biology Laboratory Sometimes that means investigating mutations in the genes that make proteins. In sickle cell anemia, for example, a mutated gene builds improperly shaped hemoglobin that sticks together and reduces the ability of red blood cells to carry oxygen. Adelman’s interest, however, lies in how otherwise normal genes are expressed—turned on or off—in the wrong amounts, at the wrong times or in the wrong tissues. In the past few years, scientists have begun to appreciate how often these instructions come from DNA segments called enhancers located far from the genes they influence. Children can be born without a pancreas when a mutation in an enhancer disrupts the “go” signal to a gene 25,000 DNA bases away that is supposed to start growing the organ. Mutations in these distant enhancers are increasingly being linked to many other diseases, including congenital heart diseases, type 2 diabetes, cancers and immunological disorders. The problem? “There’s no good way to find those enhancers,” said Adelman, professor of biological chemistry and molecular pharmacology at HMS. “If something’s wrong, we don’t know where to look.” That is now changing. Adelman and colleagues reported this week in Genes & Development that they repurposed a tool they developed in 2010, Start-seq, to generate maps of enhancers that are active in a given tissue type, disease or set of environmental conditions. Adelman believes Start-seq will help researchers seeking the sources of disrupted gene expression as well as those trying to understand how enhancers work normally. “How do enhancers give the right instructions in embryonic development and go wrong in cancer?” she said. “Not only is this stuff fascinating to explore, but we also need to answer these questions if we ever want to alter enhancers, such as to treat disease.” Already, the team has made a surprising discovery that blurs the distinction between enhancers and the genes they regulate. Who transcribes the transcribers? Like the rest of her peers, Adelman was taught in school that enhancers simply send instructions, in the form of transcription machinery, to the genes they want to “switch on.” The machines copy the genes’ DNA into RNA and use that as a blueprint to build proteins. But in 2010, researchers led by Michael Greenberg, the Nathan Marsh Pusey Professor and head of the Department of Neurobiology at HMS, discovered that enhancers in brain cells also spawn RNAs as they do their jobs—only these RNAs are tiny and short-lived, and they don’t code for proteins. Since then, the community has debated: How common is this phenomenon? What purpose, if any, do the little RNAs serve? Adelman and colleagues took advantage of the unique qualities of these RNAs to locate enhancers and get some answers. “These RNAs are very different from the ones made at genes,” Adelman explained. “They’re generated, they fall off and then they’re quickly degraded. We developed a technique to find them when they’re still stuck to the enhancers.” Rescued from the scrap heap The Start-seq technique begins with cell samples. The researchers wash away long, mature RNAs and keep ones that are still stuck to the genome. They then pluck out short RNAs that have a chemical tag characteristic of RNA-construction machinery found at genes and enhancers. Finally, the team sequences these RNAs, revealing where each came from on the genome. The result: a list of just about every enhancer that was active at the time the sample was taken, along with their exact genetic sequences. While not perfect, Start-seq returns fewer false positives and false negatives than previous enhancer-detection methods, the authors found. Poised to dive into the genetics and transcription dynamics that drive enhancers, the researchers can already answer one burning question: Around 95 percent of enhancers make RNA. “This means transcription at enhancers and protein-coding genes have much more in common than we appreciated,” said Adelman. “Philosophically it makes sense—and it helps explain why protein-coding genes can act as enhancers—but it still turns things on their head quite a bit.” The good news, she said, is that the vast knowledge scientists have gathered about control of protein-coding genes can now be applied to learning how enhancers work. Community resource The team is working to automate Start-seq so they can make it available to researchers throughout the HMS community and beyond. The tool should enable people to search for overlaps between enhancer activity and genetic variants to tease out which variants might contribute to the biological phenomenon they’re studying, whether that is Parkinson’s disease or the differentiation of stem cells. “We have plenty of neighbors who are hoping to identify relevant enhancers in their disease models,” Adelman said. “We hope to shine the flashlight on the right parts of the genome for them.” This study was supported in part by the Intramural Research Program and the National Institute of Environmental Health Sciences (Z01297ES101987) at the National Institutes of Health and by startup funds from HMS. Adelman was senior author of the paper. Telmo Henriques, a research associate in the Adelman lab, was first author.
|  Cold Spring Harbor Laboratory Press Bookstore
Cold Spring Harbor Laboratory Press Bookstore

